Development of a defacing algorithm to protect the privacy of head and neck cancer patients in publicly-accessible radiotherapy datasets
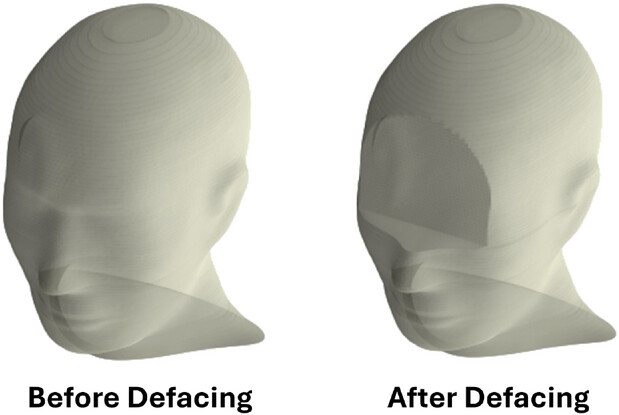 Image credit: John Kildea
Image credit: John KildeaAbstract
Background: The increase in public medical imaging datasets has raised concerns about potential patient reidentification from head CT scans. However, existing defacing algorithms, which help protect patient confidentiality, fail to preserve critical radiotherapy structures, including organs at risk (OARs) and planning target volumes (PTVs) in head and neck cancer (HNC) patients. Furthermore, current algorithms do not address the defacing of DICOM-RT structure set and dose data, which also contain information for facial surface rendering.
Purpose: To develop and validate a novel automated defacing algorithm that preserves OARs and PTVs while removing identifiable features from HNC CTs and DICOM-RT data.
Methods: Eye contours were used as landmarks to automate the removal of CT pixels above the inferior-most slice of the eye and anterior to the midpoint of the eye. Pixels within PTVs were retained if they intersected with the removed region. The body contour and dose map were then reshaped to reflect the defaced image. We validated our approach on 829 HNC CT-simulation scans from 622 patients. To evaluate privacy protection, we applied the FaceNet512 facial recognition algorithm before and after defacing on 3D-rendered CT scan pairs from 70 patients at two time points. To assess research utility, we examined the impact of defacing on auto-contouring performance using LimbusAI and analyzed the locations of PTVs relative to the defaced regions.
Results: Before defacing, the facial recognition algorithm matched 97% of patients' CT scans. After defacing, this rate dropped to just 4%. LimbusAI effectively auto-contoured organs in the defaced CTs, with perfect Dice scores of 1 for OARs below the defaced region, and mean Dice scores exceeding 0.95 for OARs on the same slices as the defaced region. PTV analysis revealed that 86% of PTVs were entirely below the cropped region, 9.1% were on the same slice as the crop without overlap, and only 4.9% extended into the cropped area. All overlapping PTVs were preserved through our algorithm’s design.
Conclusions: We developed a novel defacing algorithm that anonymizes HNC CT scans and related DICOM-RT data. Our algorithm balances patient privacy while preserving essential structures for radiotherapy research, facilitating the sharing of HNC imaging datasets for Big Data and AI.