Single-cell DNA sequencing as a means to directly examine the size and frequency of radiation-induced mutations - an exploratory study
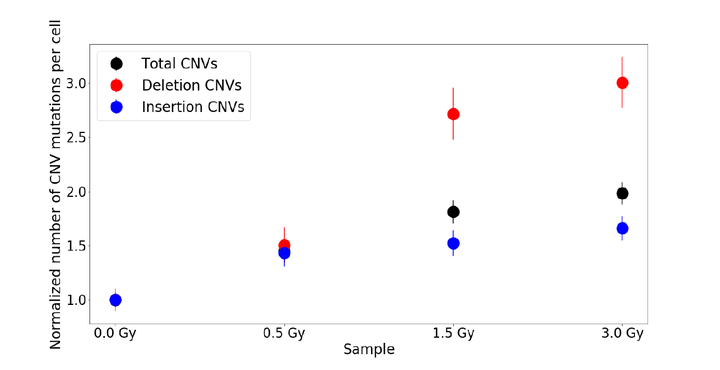 Image credit: Felix Mathew
Image credit: Felix MathewAbstract
This study examines if single-cell DNA sequencing may be used to study the mutational effects of ionizing radiation. As the action of ionizing radiation is a stochastic process, each cell in an irradiated sample experiences its own unique radiation-induced DNA damage. As a result, conventional sequencing methods such as bulk cell sequencing cannot be used to identify individual mutations. In this work, Epstein-Barr virus (EBV) transformed B-lymphoblastoid cells were irradiated with 6 MV X-ray radiation using a medical linear accelerator. Four samples of cells from the same population (400,000 cells/ml) were exposed to sham irradiation (0 Gray [Gy]; control), 0.5 Gy, 1.5 Gy and 3.0 Gy respectively at a common dose rate of about 600 cGy/min. Irradiated samples were incubated for 24 hrs and subsequently underwent single-cell whole-genome DNA sequencing to characterize the radiation impact. Mutational profiles of approximately 500 cells, randomly selected from each sample, were individually analyzed and compared to identify the variation of several mutational parameters as a function of dose. We quantified the copy number variations (CNV) for each cell in our samples. Additionally, we segregated insertion CNVs and deletion CNVs and independently analyzed their dose dependences. We found that the total number of CNVs (insertion and deletion combined) increased with dose, and the number of deletion CNVs consistently increased most. We have repeated the experiment and a new round of single-cell DNA sequencing is underway. If confirmed, our results will demonstrate that the mutational effects of ionizing radiation may be examined directly using single-cell sequencing.