 Image credit: Haley Patrick
Image credit: Haley PatrickAbstract
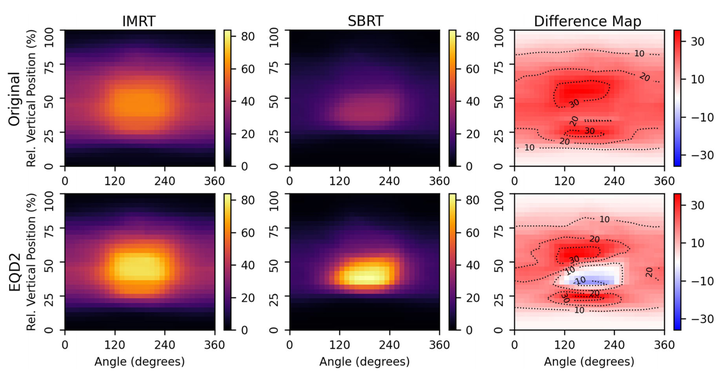
Background: Dose-outcome studies in radiation oncology have historically excluded spatial information due to dose-volume histograms being the most dominant source of dosimetric information. In recent years, dose-surface maps (DSMs) have become increasingly popular for characterization of spatial dose distributions and identification of radiosensitive subregions for hollow organs. However, methodological variations and lack of open-source, publicly offered code-sharing between research groups have limited reproducibility and wider adoption. Purpose: This paper presents rtdsm, an open-source software for DSM calculation with the intent to improve the reproducibility of and the access to DSM-based research in medical physics and radiation oncology. Methods: A literature review was conducted to identify essential functionalities and prevailing calculation approaches to guide development. The described software has been designed to calculate DSMs from DICOM data with a high degree of user customizability and to facilitate DSM feature analysis. Core functionalities include DSM calculation, equivalent dose conversions, common DSM feature extraction, and simple DSM accumulation. Results: A number of use cases were used to qualitatively and quantitatively demonstrate the use and usefulness of rtdsm. Specifically, two DSM slicing methods, planar and noncoplanar, were implemented and tested, and the effects of method choice on output DSMs were demonstrated. An example comparison of DSMs from two different treatments was used to highlight the use cases of various built-in analysis functions for equivalent dose conversion and DSM feature extraction. Conclusions: We developed and implemented rtdsm as a standalone software that provides all essential functionalities required to perform a DSM-based study. It has been made freely accessible under an open-source license on Github to encourage collaboration and community use.